※ User guide
Frequently Asked Questions:
1. Q: How to use GPS-PAIL 2.0 software?
A: You can find the latest version of GPS-PAIL 2.0 at PREDICTION page, while the stand-alone software was available at DOWNLOAD page. Currently, GPS-PAIL 2.0 webservice is implemented in php and javascript, which is compatible with various internet browsers. The stand-alone software is implemented in JAVA and could be installed on a computer with Windows/Linux/Unix/Mac OS. And we also provide a manual for users which install the package.
2. Q: What is the difference between simple prediction and comprehensive prediction?
A: The only difference between simple prediction and comprehensive prediction is that the simple prediction didn't provide annoations of surface accessbility and secondary structure. The annoations of surface accessbility and secondary structure were provided by NetSurfP ver. 1.1 (PMID: 19646261), which needs long-time computation. So, in the simple prediction, the surface accessbility and secondary structure are not visulized.
3. Q: How to read the GPS-PAIL 2.0 results?
A: Here we use the human p53 protein as the example. After clicking "Submit", the prediction results will be shown as follows:
<1>. The table of the GPS-PAIL 2.0 results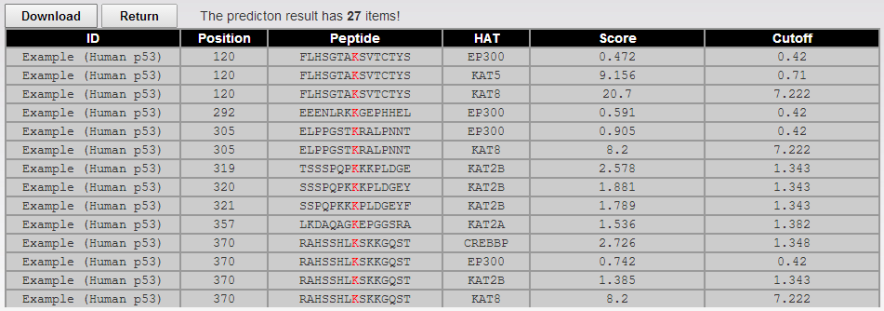
ID: The name/id of the protein sequence that you input to predict.
Position: The position of the site which is predicted to be acetylated.
Peptide: The predicted acetylation peptide with 7 amino acids upstream and 7 amino acids downstream around the modified residue.
HAT: The histone acetyltransferase which is predicted to acetylate the lysine.
Score: The value calculated by GPS algorithm to evaluate the potential of acetylation. The higher the value, the more potential the residue is acetylated.
Cutoff: The cutoff value under the threshold. Different threshold means different precision, sensitivity and specificity.
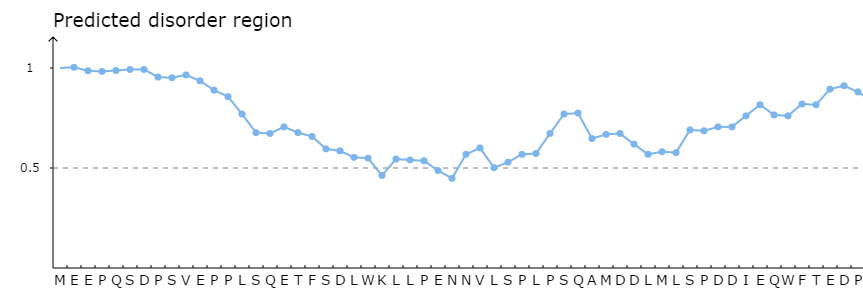
<2>. The visualization of simple prediction
Part 1: The visualization for protein disordered region predicted by IUPred [PMID: 15955779]. Cutoff = 50%, if score of prediction > cutoff, the residue is considered in disordered region.
Part 2: The visualization of the position(s) of the predicted acetylation site(s) in the protein sequence.

Part 3:
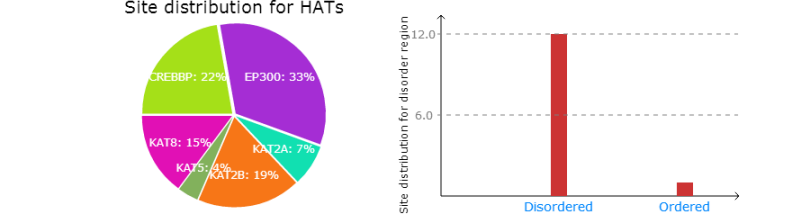
Left: The distribution of predicted acetylation sites for HATs.
Right: The distribution of predicted acetylation sites in disorder region.
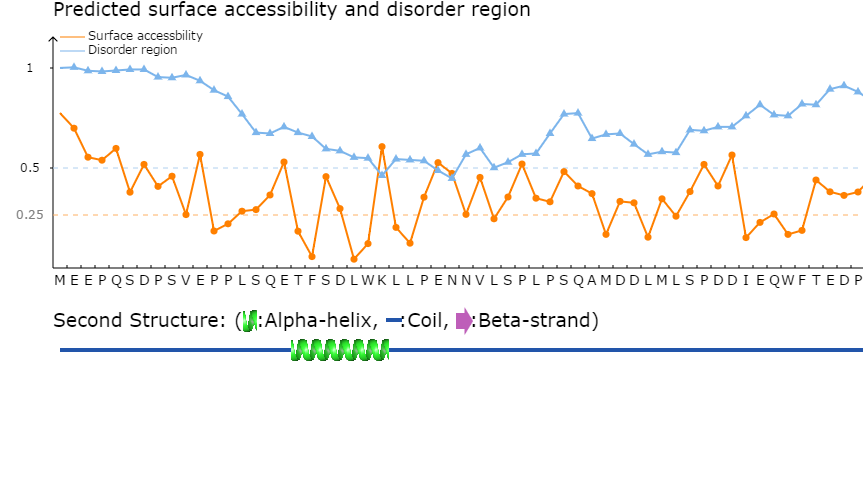
<3>. The visualization of comprehensive prediction
Part1:
Top: The surface accessbility of amino acids and the protein disordered region were predicted by NetSurfP ver. 1.1 (PMID: 19646261) and IUPred (PMID: 15955779), respectively.
The cutoff of disordered region prediction = 50%, if score of prediction > cutoff, the residue is considered in disordered region.
The cutoff of surface accessbility prediction = 25%, if score of prediction > cutoff, the residue is considered as surface exposed residue.
Bottom: The position(s) of the predicted acetylation site(s) were visualized in the protein sequence together with the secondary structure predicted by NetSurfP ver. 1.1 (PMID: 19646261).
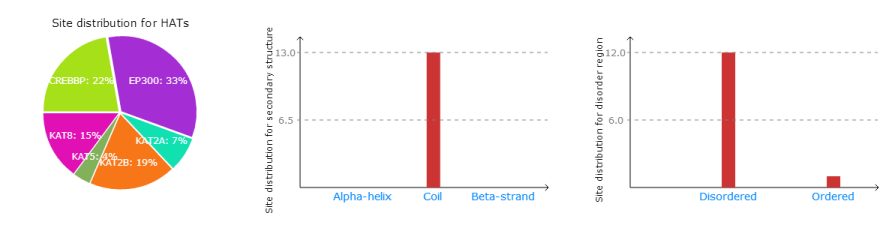
Part 2 :
Left: The distribution of predicted acetylation sites for HATs.
Middle: The distribution of predicted acetylation sites among secondary structures.
Right: The distribution of predicted acetylation sites in disorder region.
4. Q: Is GPS-PAIL 2.0 accurate? How to choose the cut-off values and the thresholds?
A: The leave-one-out (LOO) and independent testing (Testing) performance of GPS-PAIL 2.0 for 4 HATs are presented in the following table. The high threshold was suitable for large-scale prediction of acetylation sites. And the medium threshold relaxed the stringency to be useful in small-scale experiments. Also, the low threshold reduced the Sp to improve Sn considerably to be useful in exhaustively experimental identifying all potential acetylation sites in substrates.
HAT |
Positive |
Negative |
Precision |
Sensitivity |
Specificity |
|
GPS-PAIL (LOO) |
CREBBP | 167 | 1719 | 16.12% | 38.32% | 80.63% |
| EP300 | 411 | 3525 | 22.81% | 50.61% | 80.03% | |
| KAT2A | 32 | 265 | 30.56% | 68.75% | 81.13% | |
| KAT2B | 69 | 954 | 17.17% | 49.28% | 82.81% | |
| HAT1 | 10 | 110 | 62.50% | 100.00% | 94.55% | |
| KAT5 | 28 | 531 | 39.29% | 39.29% | 96.80% | |
| KAT8 | 6 | 177 | 33.33% | 83.33% | 94.35% | |
GPS-PAIL (Testing) |
CREBBP | 81 | 405 | 28.32% | 39.51% | 80.00% |
| EP300 | 85 | 605 | 25.85% | 44.71% | 81.98% | |
| KAT2A | 37 | 430 | 19.23% | 54.05% | 80.47% | |
| KAT2B | 40 | 133 | 47.92% | 57.50% | 81.20% |
5. Q: I was trying to install the software on macbook pro but my installer says the file is damaged. How can I properly install the software in Mac OS?
A: By default, Mac OS 10.8 only allows users to install applications from 'verified sources'. In effect, this means that users are unable to install most applications downloaded from the internet.
You can follow the directions below to prevent this error message from appearing.
(1) Open the Preferences. This can be done by either clicking on the System Preferences icon in the Dock or by going to Apple Menu > System Preferences.
(2) Open the Security & Privacy pane by clicking Security & Privacy.
(3) Make sure that the General section of the the Security & Privacy pane is selected. Click the icon labeled Click the lock to prevent further changes.
(4) Enter your username and password into the prompt that appears and click Unlock.
(5) Under the section labeled Allow applications downloaded from, select Anywhere. On the prompt that appears, click Allow From Anywhere.
(6) Exit System Preferences by clicking the red button in the upper left of the window. You should now be able to install applications downloaded from the internet.
6. Q: I have a few questions which are not listed above, how can I contact the authors of GPS-PAIL 2.0?
A: Please contact the authors through email, thanks.